Project safety is a crucial aspect of our project. We have implemented a series of safety measures to ensure the safety of our project and our team.
SHuffle E. coli Biosafety
We have selected the SHuffle T7-B strain as the chassis organism. The commercial SHuffle T7-B strain is derived from Escherichia coli BL21. According to the guidelines of the National Institutes of Health (NIH), the risk level of E. coli B strain is classified as Risk Group 1 (RG-1). This indicates that the B strain typically does not cause disease in healthy adults and is considered a microorganism with low individual and environmental risk.
The SHuffle strain originates from the non-pathogenic E. coli BL21 and lacks common virulence factors. Additionally, the primary pathogenic substance of E. coli, lipopolysaccharide (LPS, also known as endotoxin), cannot penetrate the intact cornea. LPS can only diffuse into the stromal layer when the cornea is damaged, and this process is mainly mediated by polymorphonuclear neutrophils (PMNs) that migrate to the damaged tissue. Therefore, no additional modification of LPS is required in this project.[2][3] Moreover, the eye is an immune-privileged site, which further reduces the risk of severe infection.
Regarding the risk of horizontal gene transfer (HGT), although engineered bacteria carrying plasmids could theoretically transfer resistance genes through conjugation, this requires specific conditions and is extremely unlikely in our application scenario.
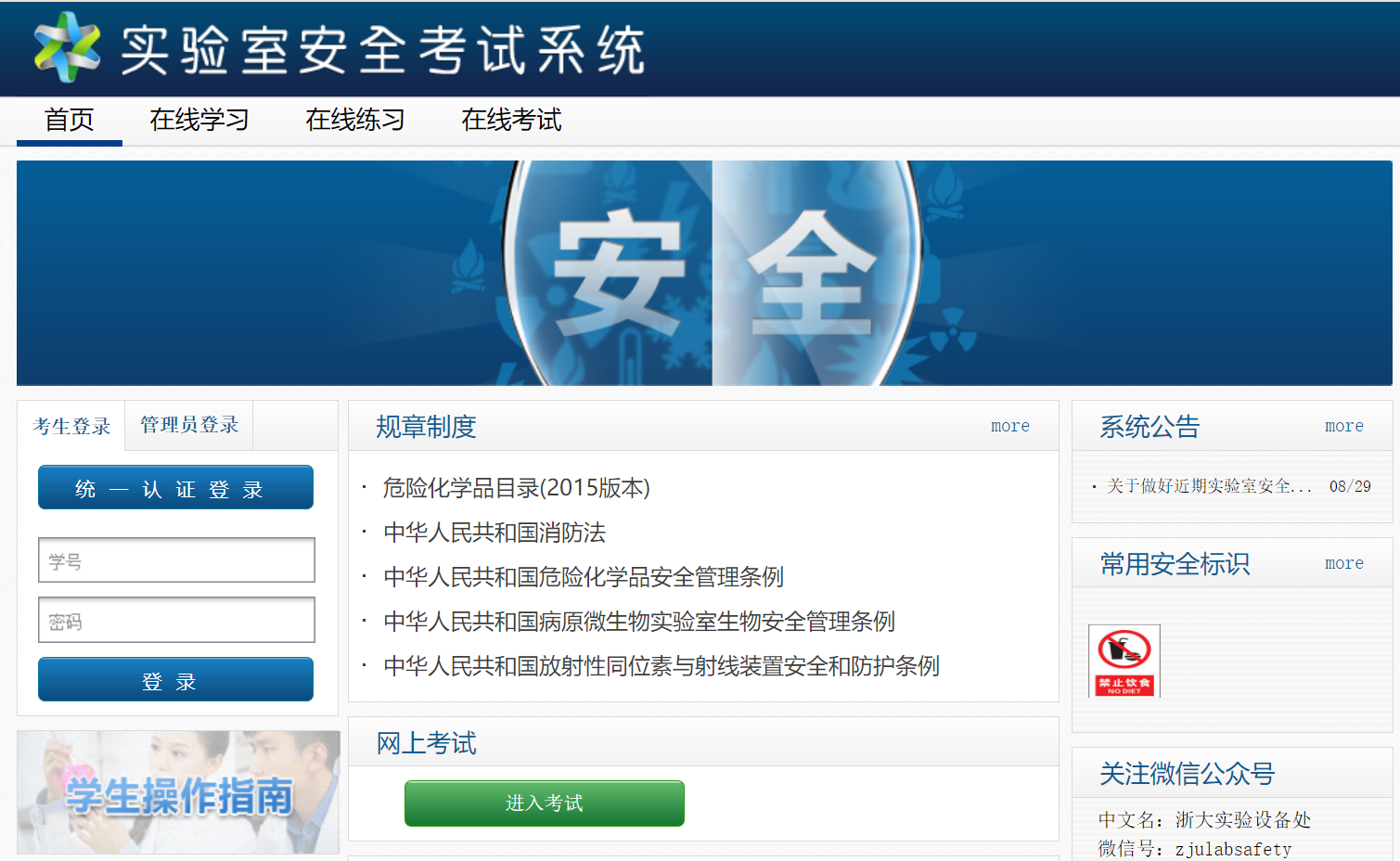
Safety Alert Module
Encapsulated in Hydrogels, the Engineered Bacteria Rely on Nutrients in Tears to Survive, and Their Viability and Reproduction Rate are Influenced by Factors such as the Ocular Microenvironment Temperature and Nutrient Elements. When the Bacterial Viability Declines and the Risk of Lysis Occurs, We Need to Promptly Generate a Reporting Signal to Alert Users to Replace the Contact Lenses. To This End, We Have Designed a Biosafety Reporting Module.
This module utilizes the fluorescent protein Crimson as the reporting signal and implements fluorescence signal detection by incorporating a light source device into the contact lens care box. The fluorescence signal intensity is proportional to the expression intensity of RNF114, and we will determine the safety grade corresponding to the fluorescence signal intensity through experiments.
The Crimson fluorescent protein is characterized by its high brightness, low toxicity, non-aggregating properties, and excellent photostability. Studies have shown that the expression of Crimson in neurons not only exhibits higher brightness than other common red fluorescent proteins (such as mCherry and mKate2), but also demonstrates significantly lower cytotoxicity during long-term expression. Moreover, Crimson does not aggregate within cells, which further reduces the potential interference with cellular functions.
By leveraging the naturally occurring polycistronic structure in prokaryotes, we achieve synchronous expression of RNF114 and the fluorescent protein Crimson. The two coding genes share a single promoter, and after translation, the two coding genes are located on the same mRNA, thereby enabling synchronous expression of the two genes.
In the polycistronic structure, the ribosome binding site (RBS) affects the reading of the open reading frame (ORF) through its complementarity to the ribosome and its distance from the start codon. We have designed RBS of varying strengths for the two coding genes to achieve relatively higher expression levels of RNF114, thereby directing translational resources primarily towards the expression of the target therapeutic molecule.
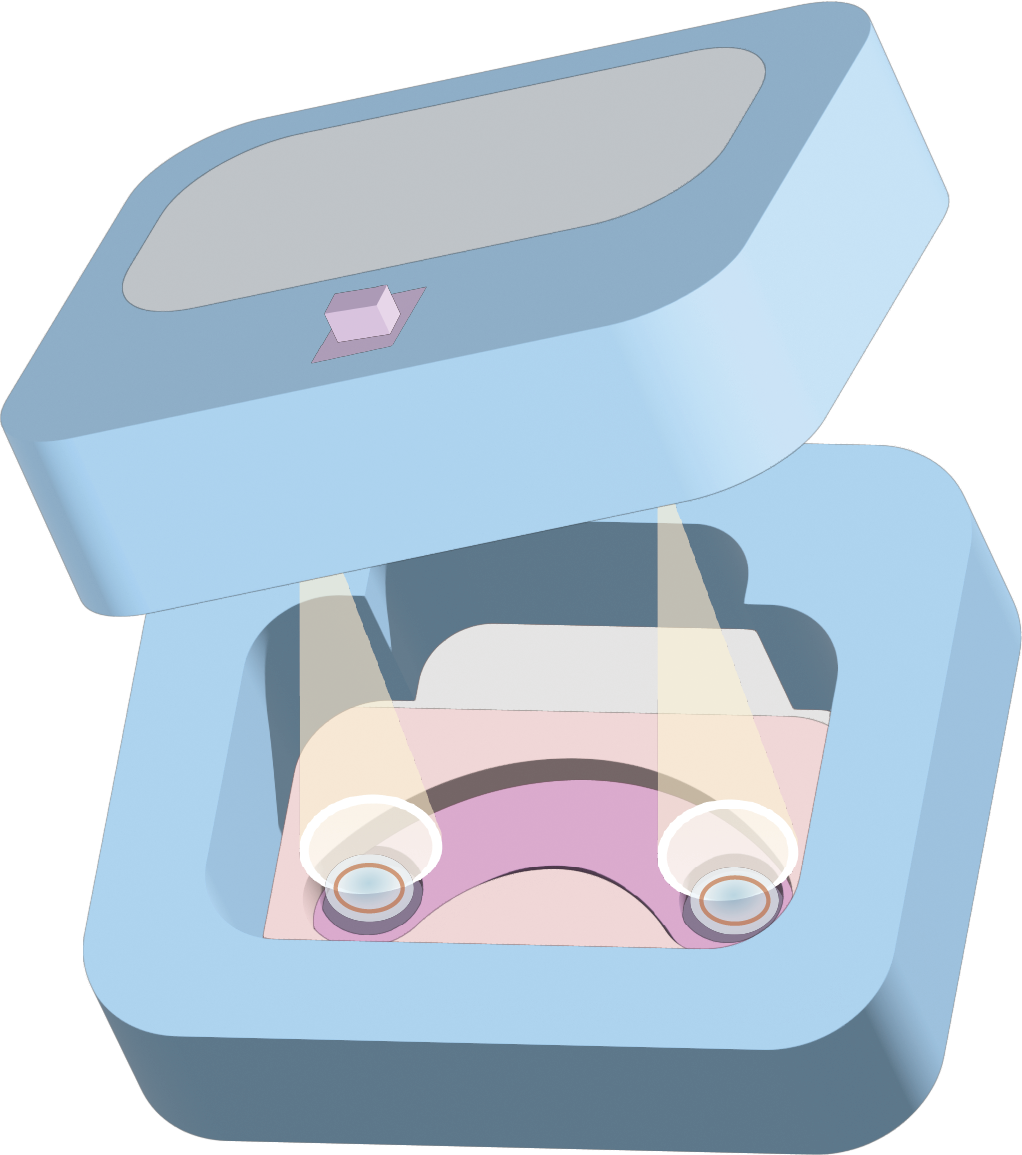
We have integrated the fluorescence signal detection device into a black contact lens care box. Upon pressing the button, the light source excites the fluorescent protein Crimson, and the fluorescence intensity emitted by the protein is detected by the BH1750 sensor. The safety grade warning is determined based on the preset threshold and displayed on the LCD screen. (See detail in Hardware Part)
Suicide Switch in case of Leakage
Nutritional Deficiency Suicide System
In this system, we use the nutrient-deficient SHuffle T7-B strain as the engineered bacterial strain and design a metabolic complementarity-based symbiotic bacterial system, ensuring that bacteria A and B can only coexist and cannot survive independently. Bacteria A cannot synthesize tryptophan (Trp) but can provide histidine (His), while bacteria B cannot synthesize histidine (His) but can provide tryptophan (Trp). These two bacteria must provide the required amino acids for each other to maintain growth; they cannot survive independently due to the lack of essential nutrients.
We knocked out the trpA gene in bacteria A to prevent it from synthesizing tryptophan, and knocked out the hisG gene in bacteria B to prevent it from synthesizing histidine. Meanwhile, bacteria A overexpress hisG to help provide histidine to bacteria B, and bacteria B overexpress trpA to help provide tryptophan to bacteria A. A strict metabolic symbiosis is formed between bacteria A and B; they can only grow in a coexistence state. If separated, they will lose their viability due to nutrient deficiencies.
This system is expected to ensure that after the failure of the hydrogel's confinement of the engineered bacteria, the bacteria will die due to the lack of essential amino acids, preventing them from spreading to the eye or the natural environment.
1. CRISPR/Cas9 knockout of genes related to trp and his synthesis
1.1 Knockout of trp synthesis-related genes in engineered strain A
1.1.1 In the trp synthesis pathway, there are four types of enzymes:
trpE: The first step in the catalytic synthesis of tryptophan is the generation of anthranilate.
trpD: Synthesis of trpC: Producing Indole-3-glycerol phosphate
trpB and trpA: Collaboratively generate the final product, tryptophan, completing the final step of tryptophan synthesis.
We chose to knock out trpC (which encodes Indole-3-glycerol phosphate synthase). Knocking out the trpC gene is expected to prevent the production of intermediates in tryptophan synthesis, thereby leading to a deficiency of tryptophan.
1.1.2 Design of gRNA
We used benchling.com to design gRNA targeting trpC. After screening multiple possible sequences, we selected:
TCGTCGCAGACAAGGCGATT
As our gRNA, its On-Target Score is 63.0, and Off-Target Score is 99.9, indicating good targeting efficiency and low off-target risk.
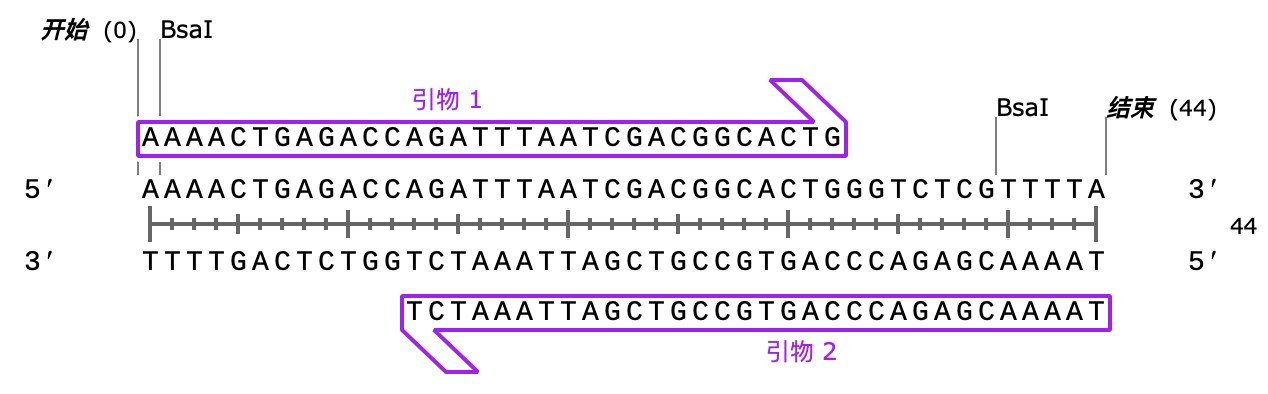
Next, we designed two primers to amplify the gRNA targeting trpC, adding Bsal restriction sites at both ends.
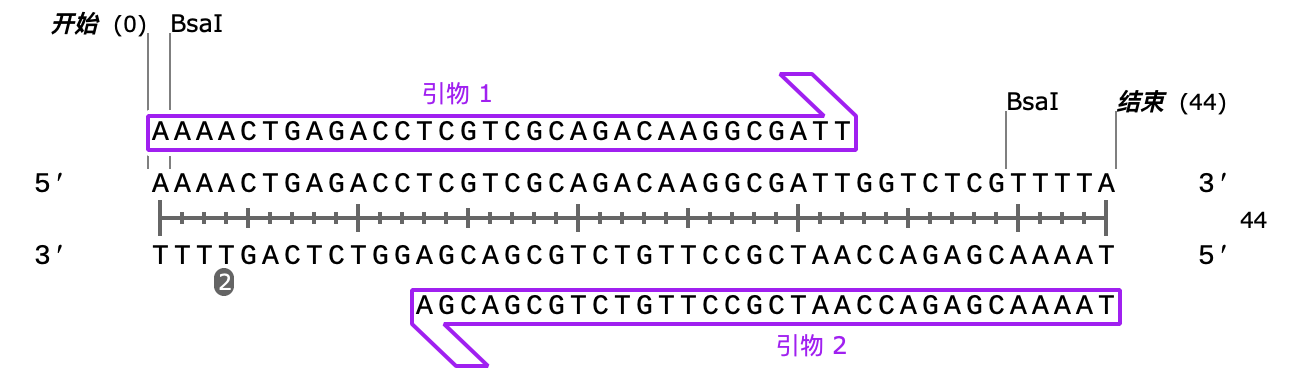
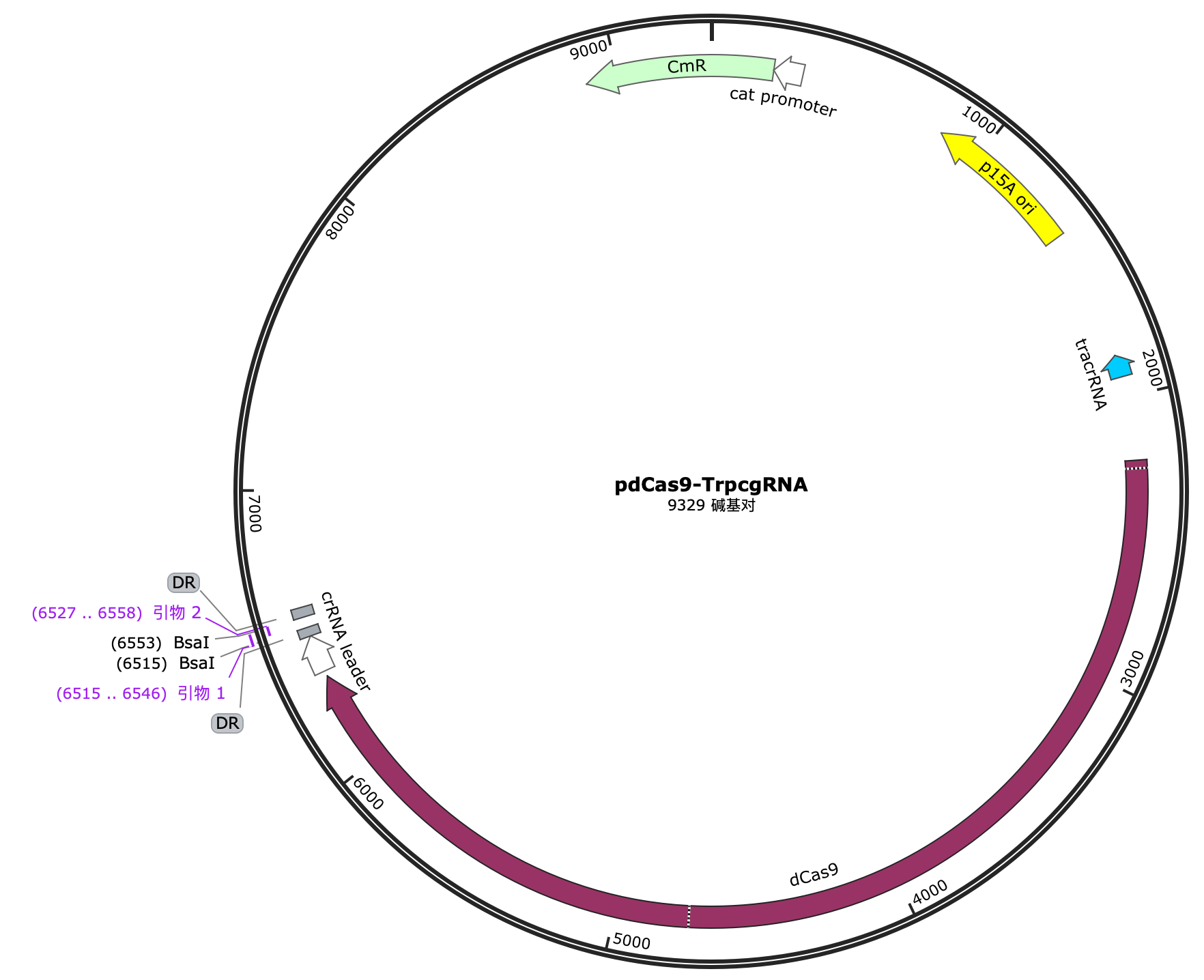
1.1.3 Construction of pdCas9-trpCgRNA plasmid
We chose pdCas9 as the vector, which has the dCas9 (dead Cas9) gene. After digestion with Bsal enzyme and ligation using T4 ligase, we obtained the pdCas9-trpCgRNA plasmid.
2. Design of the Trp Overexpression System
2.1 aroG
The aroG gene encodes 3-deamino-D-aromatic amino acid transaminase (DAHP synthase). This enzyme is a key enzyme in the aromatic amino acid synthesis pathway, involved in the synthesis of aromatic amino acids (such as tryptophan and phenylalanine). In normal metabolic processes, the activity of DAHP synthase is subject to feedback inhibition by its product (tryptophan). In other words, when the concentration of tryptophan is too high, it inhibits the activity of the enzyme encoded by aroG, thereby reducing the synthesis of aromatic amino acids.
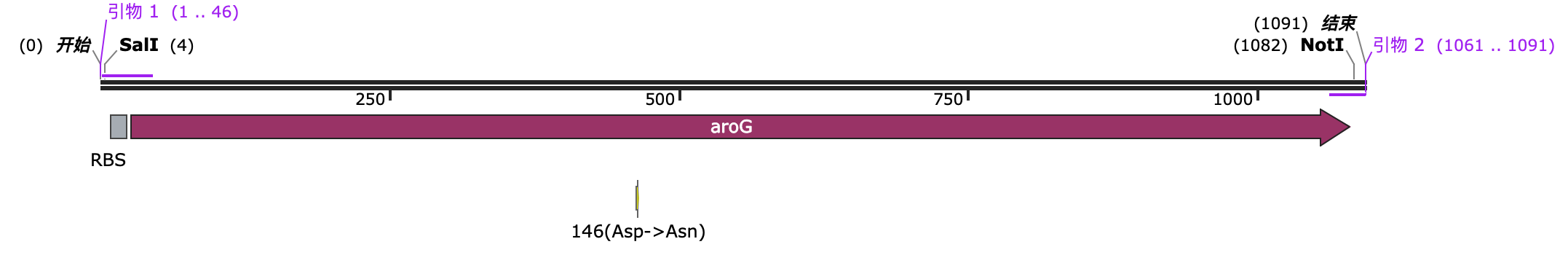
We modified the aroG gene by mutating the aspartic acid (Asp) at position 146 to asparagine (Asn) (i.e., D146N mutation). This removed the feedback inhibition of tryptophan on the enzyme. The enzyme's activity is no longer inhibited by high concentrations of tryptophan, so even if there is a high concentration of tryptophan in the cells, DAHP synthase can still continue to catalyze the synthesis of aromatic amino acids, avoiding metabolic pathway blockage due to feedback inhibition.
We added an RBS to the front of its CDS sequence to facilitate the formation of a polycistronic transcript with the enzymes related to Trp synthesis, allowing for simultaneous expression. We also added SalI and NotI restriction enzyme recognition sites at both ends to facilitate insertion into a subvector.
2.2 Knockout of His Synthesis Related Genes in Engineering Strain B
2.2.1 In the His synthesis pathway, there are five types of enzymes:
- hisG: Catalyze Imidazoleglycerol-phosphate dehydratase to produce Imidazoleglycerol.
- hisD: Synthesize Imidazoleacetol.
- hisB: Synthesize Histidinol.
- hisA: Synthesize Histidinaldehyde.
- hisC: Synthesize Histidine (final product).
We chose to knock out the hisA gene, which encodes Phosphoribosyl-ATP synthase. In the histidine synthesis pathway, hisA is a key enzyme responsible for generating Phosphoribosyl-ATP. By knocking out the hisA gene, we can block a critical step in histidine synthesis, thereby preventing the synthesis of histidine.
2.2.2 Design of gRNA
We used benchling.com to design gRNA targeting hisA. After screening multiple possible sequences, we selected:
TCGTCGCAGACAAGGCGATT
As our gRNA, its On-Target Score is 72.0, and Off-Target Score is 99.8, indicating good targeting efficiency and low off-target risk.
Next, we designed two primers to amplify the gRNA targeting hisA, adding Bsal restriction sites at both ends.
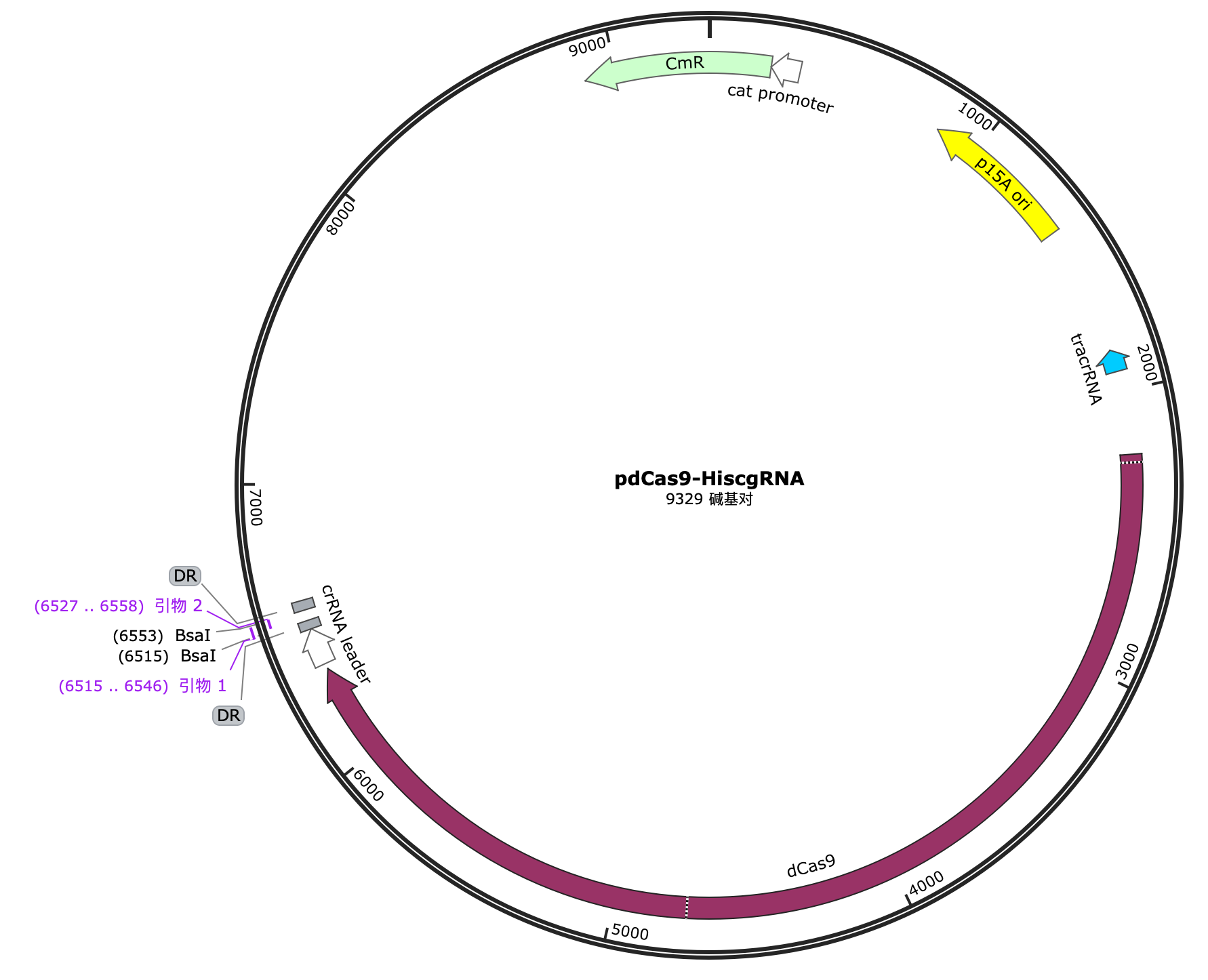
2.2.3 Construction of pdCas9-hisAgRNA plasmid
We selected pdCas9 as the vector, which has the dCas9 (dead Cas9) gene. After digestion with Bsal enzyme and ligation using T4 ligase, we obtained the pdCas9-hisAgRNA plasmid.
Phosphate Killing Switch
If nutrient deficiency suicide systems significantly affect the expression of fusion proteins, we plan to adopt a killing switch strategy. We use SHuffle T7-B as the engineered strain and have designed a bacterial suicide system based on phosphate concentration to ensure that the bacteria can only survive within the hydrogel encapsulated environment and cannot survive independently.
1. Suicide Switch Design
The eyes are very sensitive organs, and inappropriate molecules can cause irritation to the eye tissue, leading to burning sensations, tingling, itching, the feeling of foreign bodies, or tearing. It can even cause severe consequences such as corneal damage. Therefore, traditional operons such as arabinose and glucose are not suitable for this system's Killing Switch design.
Phosphate is commonly used in emergency eyewash solutions to help neutralize chemical damage to the eyes and regulate the osmotic pressure of the rinse solution. Thus, it is an ideal suicide switch.
We have drawn inspiration from the 2021-SZU-China team, who designed a phosphate suicide switch. We use the phosphate-sensitive promoter pPhoB, but unlike that project, the expression of the suicide gene is inhibited at normal phosphate concentrations and promoted at low phosphate concentrations. When the PhoB promoter is activated, transcription occurs, and the suicide gene is expressed. If the engineered bacteria accidentally leak or leave the hydrogel environment, they will be killed.
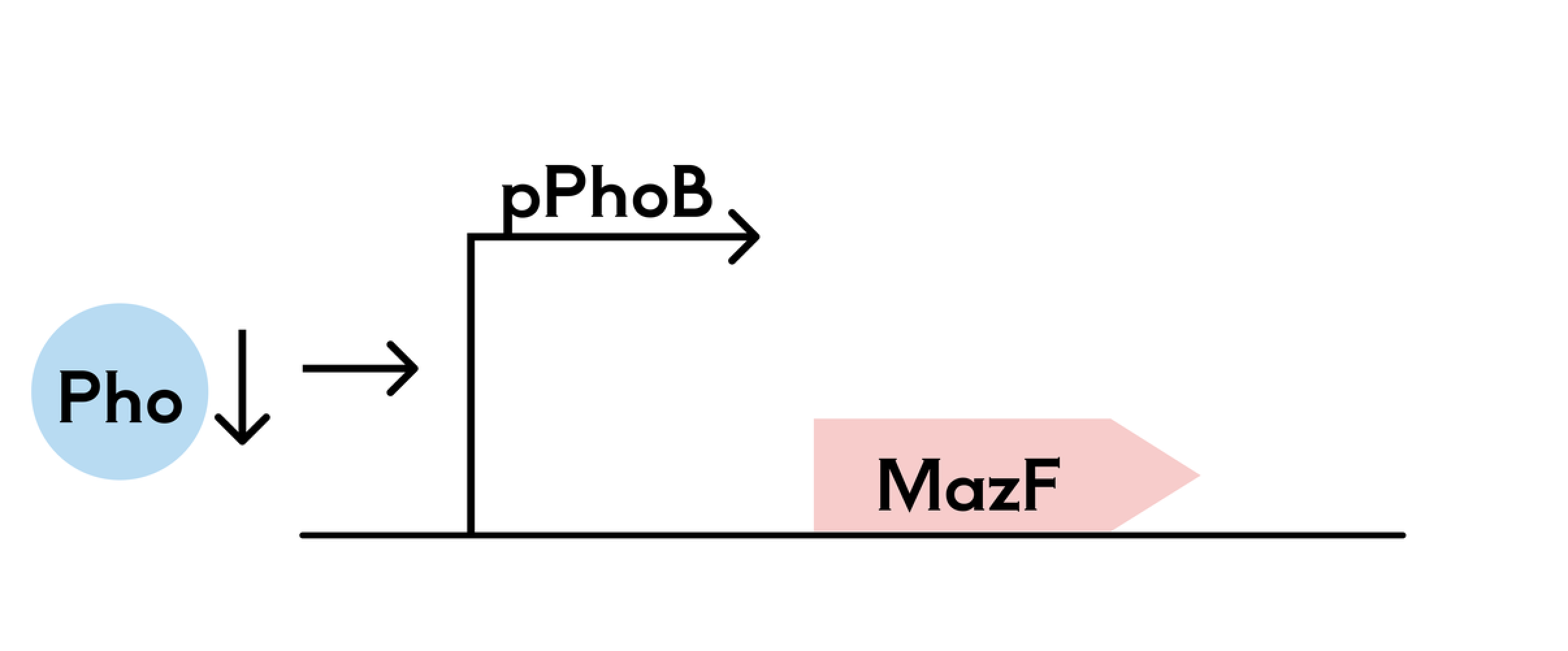
2. pET-32a(+)-PHOB-mazF Plasmid Construction
We selected the pET-32a(+) plasmid as a vector, and through PCR, we connected the recognition sequences for the restriction enzymes SgrAI and XbaI at both ends of the phosphate-sensitive promoter PHOB. Similarly, we connected the recognition sequences for BamHI and HindIII at both ends of the suicide gene mazF. After double restriction enzyme digestion, the fragments were inserted into pET-32a(+).
-PHOB-mazF.png)
3. Hydrogel Sustained Release Design
To maintain a high phosphate concentration around the engineered bacteria, we chose Poly(2-hydroxyethyl methacrylate - methacrylate copolymer - hyaluronic acid - β-cyclodextrin) hydrogel for loading. Since phosphate carries a negative charge in the hydrogel environment, the long-chain hyaluronic acid (HA), which also has a negative charge, can strongly restrict its diffusion. Additionally, the β-cyclodextrin in the hydrogel composition has a hydrophobic cavity and a hydrophilic outer surface, making it an effective storage material for phosphate.
As for the phosphate concentration in the hydrogel, data shows that the phosphate concentration in the eye is typically similar to that in physiological fluids, around 0.8–1.5 mmol/L. Phosphate concentrations in soil vary widely, typically ranging from 0.001–0.1 mmol/L. In natural water bodies, phosphate concentrations are generally low, around 1.05 × 10⁻⁷ - 5.25 × 10⁻⁷ mol/L (corresponding to 0.01–0.05 mg/L P). In eutrophic water sources, the phosphate concentration is roughly between 8.34 × 10⁻⁷ mol/L and 8.34 × 10⁻⁶ mol/L.
We aim to load the hydrogel with a phosphate concentration higher than all the concentrations mentioned above, while ensuring it does not irritate the eye, even considering the dilution effect of tears.
Using the above formula, we designed the phosphate concentration in the hydrogel to be 1.82mmol/L
Production
The engineered bacteria will be produced in laboratories that comply with biosafety standards. Both the cultivation of the engineered bacteria and the production process of the contact lenses will adhere to international biosafety standards. The cultivation of Escherichia coli must be conducted in a controlled environment to minimize the risk of leakage. Operators must be specially trained and strictly follow operating procedures, including the use of personal protective equipment and biosafety cabinets. Waste must be treated according to the standards for biohazardous waste to prevent the leakage of biological materials.
Transportation
After the engineered bacteria are encapsulated in the hydrogel, we will package the contact lenses in sterile, sealed bags for distribution. During transportation and storage, we plan to use a cold chain at 4°C to ensure that the engineered bacteria remain highly viable and capable of reproduction when they reach the end-user. We will conduct experimental tests to determine and clearly label an expiration date for the product. Given the unique nature of live bacterial products, we aim to achieve a just-in-time production and usage model to the greatest extent possible.
Use
We provide users with a reusable contact lens care box, which allows users to place their contact lenses into the box when they are not wearing them. The box is equipped with a fluorescence detection function that can assess the safety and usability of the contact lenses. Users can determine whether they need to replace their contact lenses based on the safety grade displayed on the LCD screen.
We will also provide instructions guiding users on how to disinfect expired contact lenses with 75% ethanol or other disinfectants before placing them into a sealed bag for disposal. This procedure is designed to minimize the potential leakage of engineered bacteria to the greatest extent possible.